As a precursor to my work on full 3D reconstructions of neural mitochondria I developed a method to automatically segment mitochondria from individual image slices. Our approach improved over previous attempts at segmenting neural EM imagery using normalized cuts, level sets, textons, and active contour approaches and a convolutional neural network/watershed.
This is because the image cues other methods rely on are not discriminative enough. Mitochondria exhibit irregular shapes not easily captured and their texture can easily be confused with that of groups of vesicles or other organelles. Mitochondrial boundaries are difficult to distinguish from other membranes that share a similar appearance. Overcoming these difficulties requires taking all visible image cues into account simultaneously.
Our approach overcomes these issues by
- Using all available image cues simultaneously. We consider powerful shape cues that do not require an explicit shape model in addition to texture and boundary cues.
- Learning the appearance of mitochondrial membranes. We train a classifier to predict where mitochondrial boundaries occur on a superpixel graph using these cues.
We first produce a superpixel oversegmentation of the image to reduce computational cost and enforce local consistency. The superpixels define nodes in a graph used for segmentation. We then extract sophisticated shape, texture, and boundary cues captured by Ray and Rotational features for each superpixel. Support vector machine (SVM) classifiers are trained on these features to recognize the appearance of superpixels belonging to mitochondria, as well as pairs of superpixels containing a mitochondrial membrane. Classification results are converted to probabilities, which are used in the unary and pairwise terms of a graph-cut algorithm that segments the superpixel graph. Finally, a postprocessing step smooths the segmentation. We show qualitatively and quantitatively that our approach yields substantial improvements over existing methods.
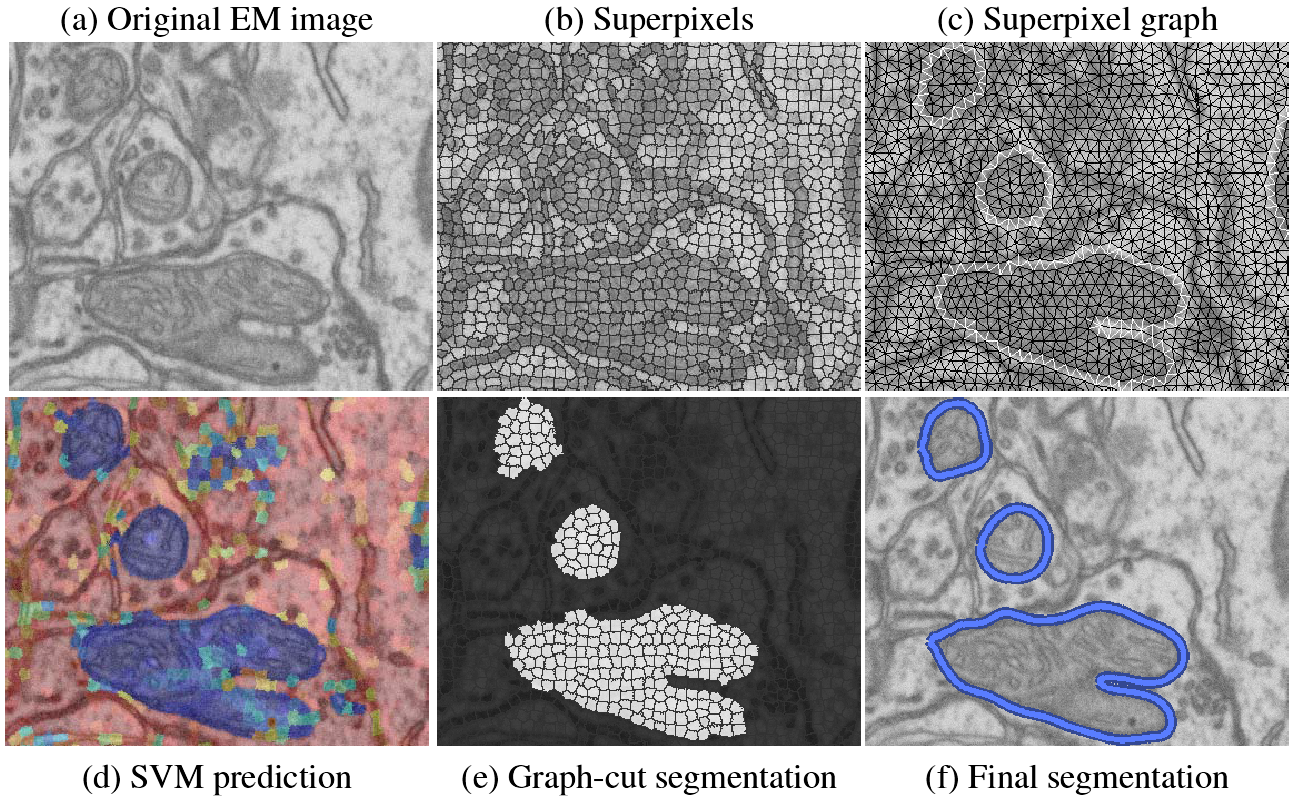
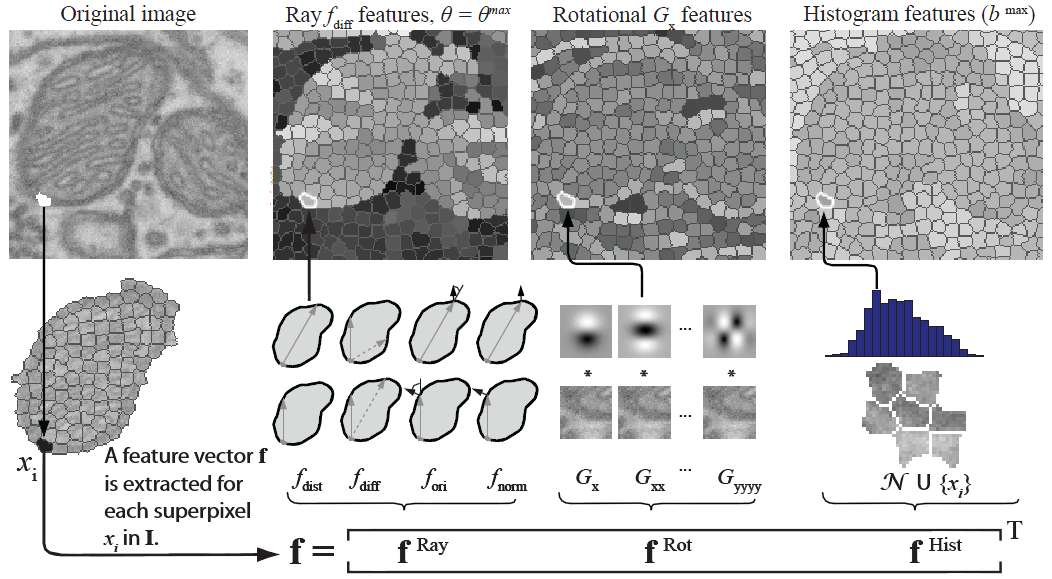
We use SVMs to predict which superpixels contain mitochondria and which neighboring superpixels contain a mitochondrial boundary. Robust shape, texture, and boundary cues are all essential to this process and must be provided to the SVM. For every superpixel, we extract the following image cues: a ray descriptor that captures object shape, rotational features describing texture and boundaries, and intensity histograms describing the intensity profile.
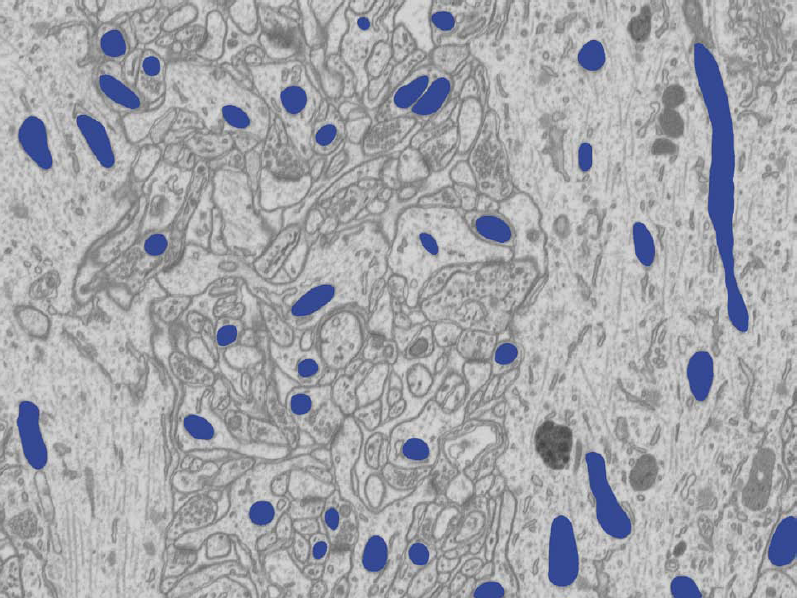
Our experiments on several EM slices demonstrate that our approach significantly outperforms state-of-the-art techniques including TextonBoost and Fulkerson, and closely matches the performance of human annotators.




References
- A. Lucchi, K. Smith, R. Achanta, V. Lepetit, and P. Fua, A Fully Automated Approach to Segmentation of Irregularly Shaped Cellular Structures in EM Images, International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), Beijing, China, September 2010